There is plenty of evidence showing that autophagy plays an important role in the biological process of cancer. The purpose of this study was to establish a novel autophagy-related prognostic marker for lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC).
Methods
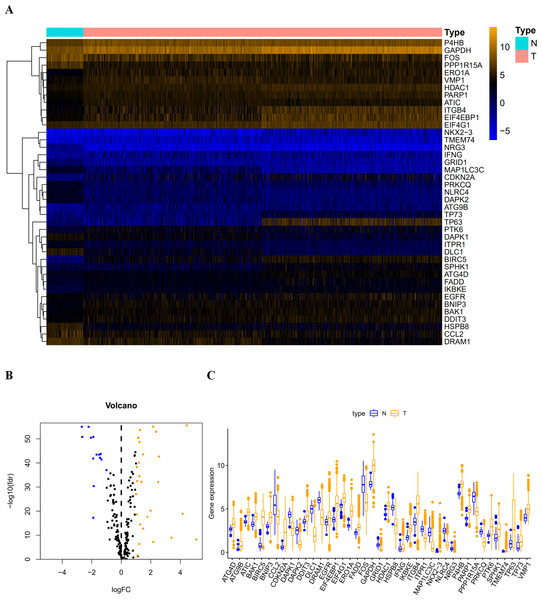
The mRNA microarray and clinical data in The Cancer Genome Atlas (TCGA) were analyzed by using a univariate Cox proportional regression model to select candidate autophagy-related prognostic genes. Bioinformatics analysis of gene function using the Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) platforms was performed. A multivariate Cox proportional regression model helped to develop a prognostic signature from the pool of candidate genes. On the basis of this prognostic signature, we could divide LUAD and LUSC patients into high-risk and low-risk groups. Further survival analysis demonstrated that high-risk patients had significantly shorter disease-free survival (DFS) than low-risk patients. The signature which contains six autophagy-related genes (EIF4EBP1, TP63, BNIP3, ATIC, ERO1A and FADD) showed good performance for predicting the survival of LUAD and LUSC patients by having a better Area Under Curves (AUC) than other clinical parameters. Its efficacy was also validated by data from the Gene Expression Omnibus (GEO) database.
Conclusion
Collectively, the prognostic signature we proposed is a promising biomarker for monitoring the outcomes of LUAD and LUSC.

Δεν υπάρχουν σχόλια:
Δημοσίευση σχολίου